


Our studies focus on the mechanisms by which organisms sense their surroundings and adapt appropriately. Those properties are particularly important for pathogenic bacteria, which produce virulence-associated factors at specific times and locations during infectious processes. Understanding those regulatory mechanisms can identify key targets for novel therapies to treat and prevent infectious diseases.
The Lyme disease spirochete, Borrelia burgdorferi, is an especially useful model for studies of gene and protein regulation in a pathogen. In addition to being the cause of a significant human disease, B. burgdorferi infects both vertebrates and ticks. The Lyme spirochete must, therefore, possess mechanisms to accurately determine which of those two hosts it is in, and produce proteins and other factors that are essential for each stage of its infectious cycle. Plus, B. burgdorferi must recognize when a tick is feeding, in order to undergo the physiological changes that are necessary for transmission into the vertebrate host. Our long term goal is to identify ways to block signaling in the Lyme spirochete, thereby "confusing" the bacteria so that they do not produce critical virulence factors and make them easier to clear from human and other vertebrate hosts.
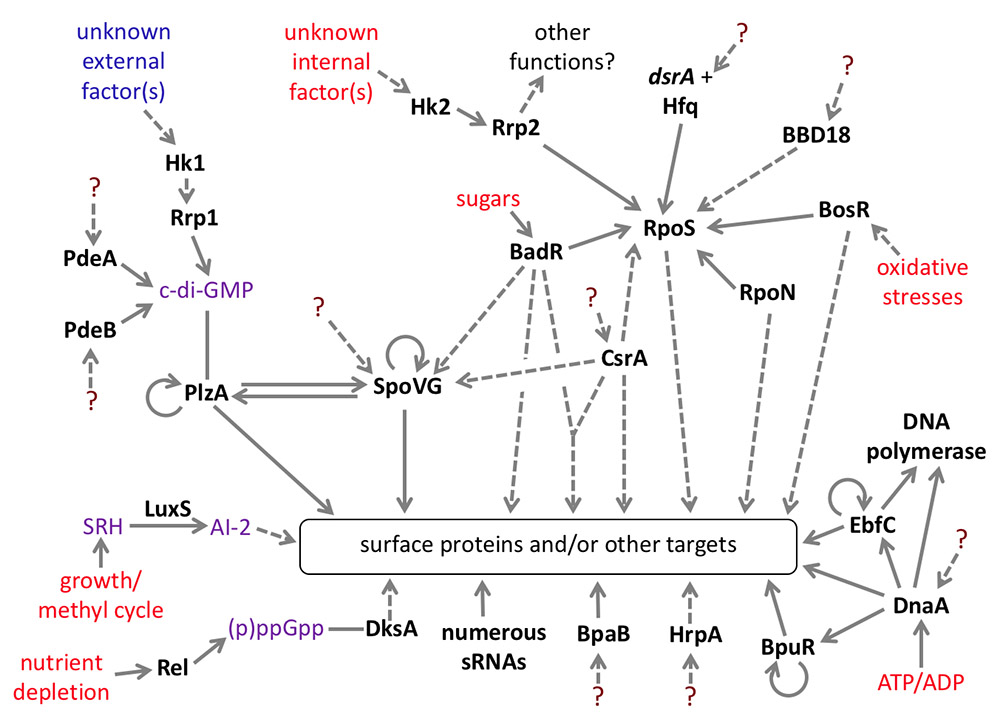
Data from our lab and others indicate that B. burgdorferi controls levels of virulence-associated proteins through a network of DNA- and RNA-binding proteins, small RNAs, modified nucleotides, and other molecules. All evidence indicates that each bacterial operon is controlled through interplay of multiple, different factors.
Our early studies primarily focused on the mechanisms by which B. burgdorferi controls expression of its erp operons, and the functions of the Erp outer surface proteins. We and others determined that Erp proteins interact with vertebrate host components, including plasmin, laminin, glycosaminoglycans, and complement factors H, C1s, and C1r. Production of Erp proteins is induced when vector ticks begin feeding on vertebrate hosts, and are produced throughout vertebrate infection. Our investigations demonstrated that transcription of erp operons is controlled by the BpaB repressor, the EbfC antirepressor, and the BpuR co-repressor. Moreover, each known regulatory factor appears to affect several different operons, and may do so in different ways. For example, the BpaB protein represses erp transcription, yet enhances transcription of the cp32 prophage ssbP (single-stranded DNA-binding protein) and nucP (nuclease) genes.
Those investigations have expanded to include additional B. burgdorferi infection-associated proteins. We recently found that a protein named Gac is a transcriptional repressor of the essential virulence factor OspC.
Over the years, our lab has identified several novel DNA- and RNA- binding proteins of B. burgdorferi. Among these are EbfC, which we demonstrated to be both a transcriptional regulatory factor and a novel type of nucleoid-associated protein. All species of Eubacteria encode a homologue of EbfC, although their function was not known prior to our studies. The BpuR protein is homologous to the PUR proteins of higher eukaryotes, so studies of the borrelial protein are providing insights on regulatory mechanisms of humans. Firmicutes, some spirochetes, and members of other taxa produce a SpoVG protein, which we demonstrated to be a nucleic acid-binding protein.
Currently, we are working on four major topics:
PlzA: This is the only B. burgdorferi protein that binds the messenger molecule cyclic-di-GMP (c-di-GMP). PlzA bound with c-di-GMP binds to specific DNA sequences in the borrelial genome, while un-liganded PlzA cannot bind DNA. We are defining the functions of PlzA and c-di-GMP in regulation of protein expression during infection processes. Supported by NIH R01 AI144126.
OspC: An outer surface lipoprotein, OspC is required for mammalian infection. Production of OspC by B. burgdorferi is induced during feeding of colonized ticks on vertebrate hosts. Our investigations of the mechanisms by which the Lyme spirochete controls OspC production have, to date, identified and characterized three DNA-binding proteins and a non-coding RNA.
BpuR: Another small RNA-binding protein that controls translation of critical B. burgdorferi proteins. Supported by NIH R21 AI139956.
DnaA: We found that B. burgdorferi controls production of numerous virulence factors in response to changes in bacterial replication rate. We then linked that to control of regulatory factors through DnaA, the master regulator of chromosomal replication. Supported by NIH R21 AI147139.

brian.stevenson@uky.edu
Ph.D. from Stony Brook University
Postdoctoral studies at Yale University and Rocky Mountain Laboratories, N.I.H.
 The Stevenson Lab on Blue Sky
The Stevenson Lab on Blue Sky
 The Stevenson Lab on Twitter
The Stevenson Lab on Twitter
 The Stevenson Lab on Facebook
The Stevenson Lab on Facebook